- Proteomes NCBI & Phytozome
- Proteomes UniProt
Search Proteome Virtual Map
Instructions for Proteome Search
- Select Database Proteomes NCBI & Phytozome or Proteomes UniProt
- Select Species from drop-down list or Search by Species name Keyword.
- After selecting Specie, 2D virtual Proteome Map will display. Sample data of Molecular Weight and Isoelectric point from which 2D Map generated will Display.
- If required, you can download 2D map by clicking on download button and complete file containing Molecular Weight & Isoelectric point by clicking download Link.
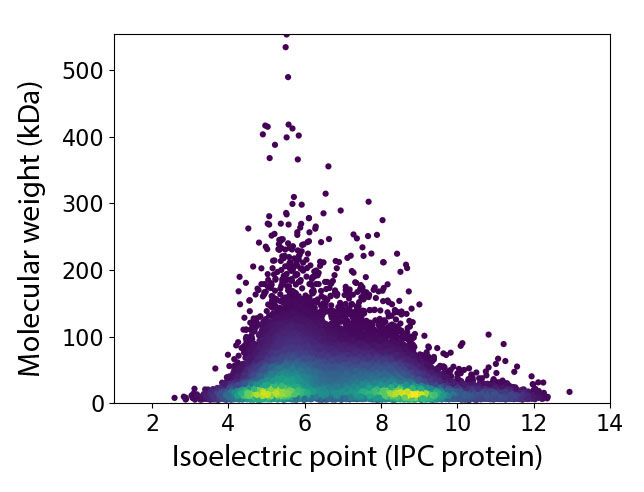
Virtual 2-D Proteome Map
Species Name: Aegilops tauschii
'; Download 2D Map
Download 2D MapDownload complete file of Molecular weight and Isoelectric point from which 2D Map of Proteome Generated.(Aegilops tauschii)
Molecular Weight used for Proteome Map
Sample data is given below if you want to download complete data click on above link below the 2-D map
Aegilops tauschii
| Sr No. | Accession No. | Protein Name | Mol. Wt (kDa) | pI |
|---|---|---|---|---|
| 1 | XP_020146151.1 | uncharacterized protein At4g37920, chloroplastic | 48.09 | 5.3 |
| 2 | XP_020146153.1 | uncharacterized protein LOC109731410 | 14.3 | 5.7 |
| 3 | XP_020146154.1 | uncharacterized protein LOC109731412 | 26.8 | 10.2 |
| 4 | XP_020146165.1 | uncharacterized protein LOC109731427 | 36.5 | 5.0 |
| 5 | XP_020146167.1 | uncharacterized protein LOC109731430 | 17.8 | 4.4 |
| Sr No. | Accession No. | Protein Name | Mol. Wt (kDa) | pI |
Search Proteome Virtual Map
Instructions for Proteome Search
- Select Database Proteomes NCBI & Phytozome or Proteomes UniProt
- Select Species from drop-down list or Search by Species name Keyword.
- After selecting Specie, 2D virtual Proteome Map will display. Sample data of Molecular Weight and Isoelectric point from which 2D Map generated will Display.
- If required, you can download 2D map by clicking on download button and complete file containing Molecular Weight & Isoelectric point by clicking download Link.
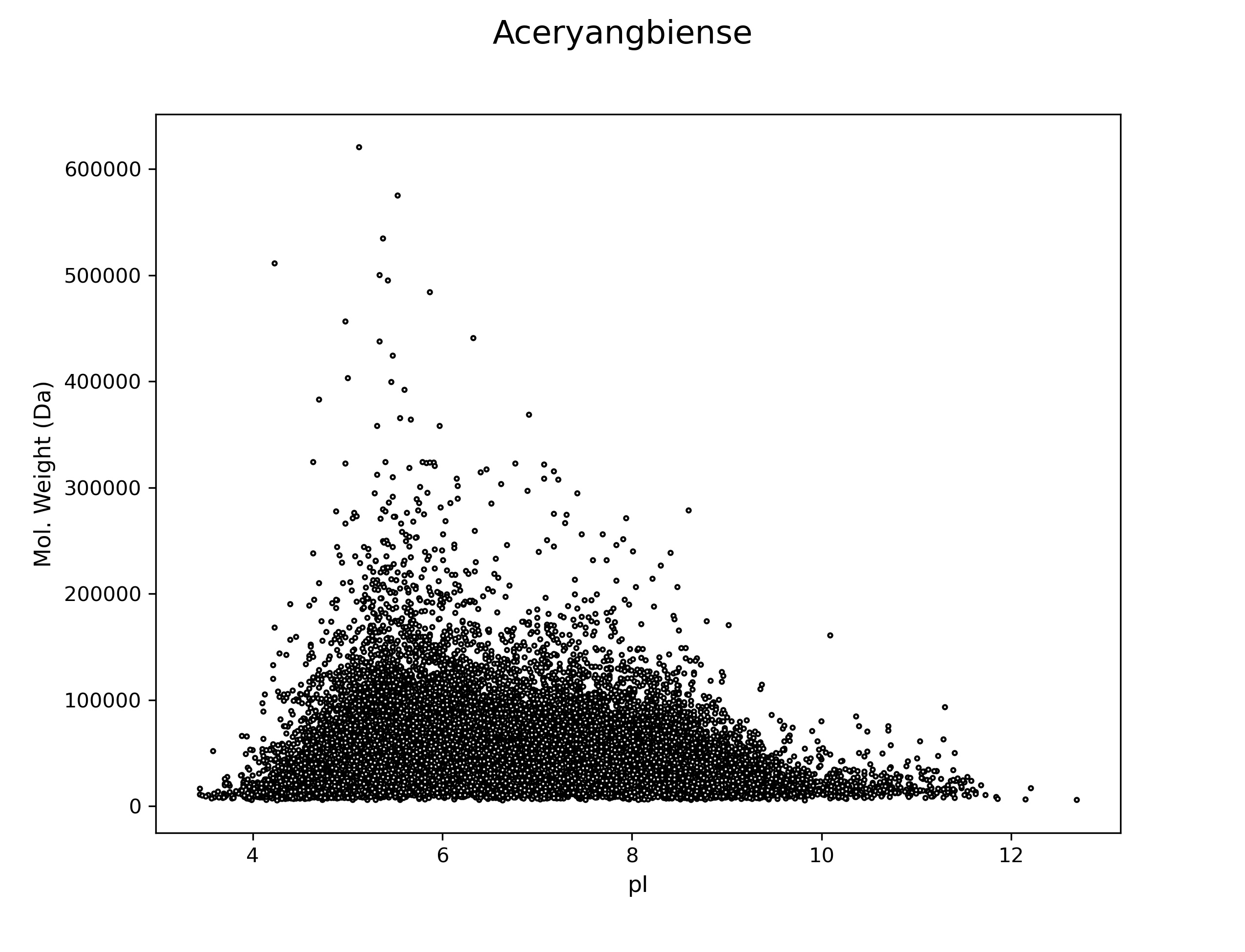
Virtual 2-D Proteome Map
Molecular Weight used for Proteome Map
Sample data is given below if you want to download complete data click on above link below the 2-D map
Acer yangbiense
| Sr No. | Accession No. | Protein Name | Mol. Wt (kDa) | pI |
|---|---|---|---|---|
| 1 | A0A5C7GMY8 | A0A5C7GMY8_9ROSI Uncharacterized protein OS=Acer yangbiense OX=1000413 GN=EZV62_028320 PE=4 SV=1 | 24.7 | 9.21 |
| 2 | A0A5C7GN06 | A0A5C7GN06_9ROSI Abhydrolase_3 domain-containing protein OS=Acer yangbiense OX=1000413 GN=EZV62_028312 PE=4 SV=1 | 51.76 | 7.45 |
| 3 | A0A5C7GN15 | A0A5C7GN15_9ROSI Protein N-terminal glutamine amidohydrolase OS=Acer yangbiense OX=1000413 GN=EZV62_028296 PE=3 SV=1 | 19.78 | 5.94 |
| 4 | A0A5C7GN26 | A0A5C7GN26_9ROSI Uncharacterized protein OS=Acer yangbiense OX=1000413 GN=EZV62_027763 PE=4 SV=1 | 18.28 | 7.82 |
| 5 | A0A5C7GN35 | A0A5C7GN35_9ROSI Uncharacterized protein OS=Acer yangbiense OX=1000413 GN=EZV62_028318 PE=4 SV=1 | 18.94 | 4.8 |
| Sr No. | Accession No. | Protein Name | Mol. Wt (kDa) | pI |